Elisa BOMBARDA: Research Interests
My major scientific interests focus on the investigation of the thermodynamics and the kinetics of the interactions of proteins as well as of their conformational dynamics using fluorescence spectroscopy and other biophysical techniques. The goal of my research activity is to provide a detailed description of the involved molecular mechanisms in order to understand and predict their biological implications.
SCIENTIFIC TECHNIQUES
- Steady-state fluorescence spectroscopy
- Time resolved fluorescence spectroscopy
- UV-Vis Absorption spectroscopy
- Fast kinetics - Stop flow technique
- Potentiometry
- Determination of protonation titration curve by 1D and 2D 1H NMR
- Theoretical methods (Continuum electrostatics and Molecular mechanics)
TOPICS
Theoretical investigations on Phytochelatin synthase (PCS).In order to understand the binding of GSH to PCS, a series of electrostatic and molecular mechanic calculations were performed. This modeling together with a series of electrostatic calculations indicate that electrostatic interactions play a crucial role in the binding of GSH to PCS. The binding pocket for GSH on PCS is rather positively charged (Figure 1), which makes it suitable to bind the overall negatively charged GSH molecule. These observations have consequences for the labeling of GSH in the planned fluorescence studies.
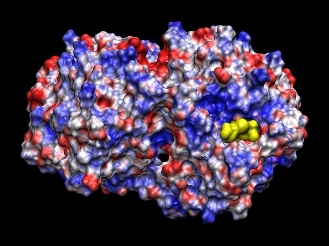
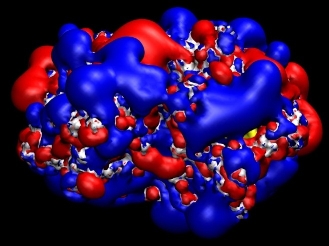
Theoretical analysis of protonation in proteins.
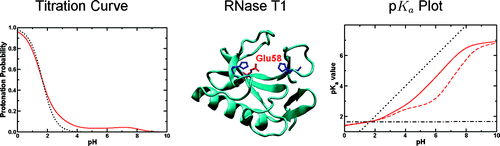
The protonation of a protein determines its electrostatics, thus it is important to rationalize its interactions with other molecules. Moreover, the knowledge of protonation energy is often important to describe enzymatic mechanisms or proton transfer reaction. With a pure mathematical approach connected to electrostatic calculations we clarify the meaning of titration curves of residues in proteins and how they are connected to pK values and protonation free energy. We show that there are at least two different meaningful ways of deriving pK values from titration curves. One relates to thermodynamic data and the other to kinetic data. pK values derived from thermodynamic measurements and kinetic measurements may have different meanings depending on whether the protonation can equilibrate during the reaction or not. We could show under which conditions the two ways are equivalent.
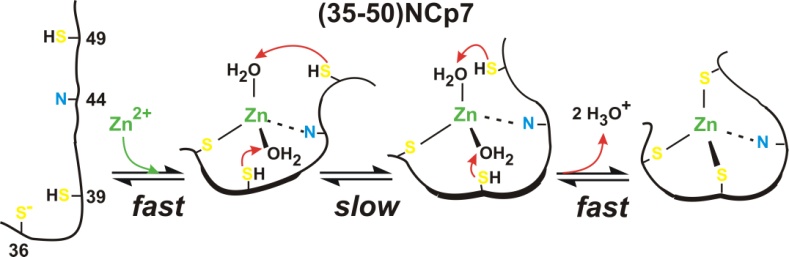
Metal ion binding to cysteine-containing peptides using fluorescence, UV-Vis, and NMR spectroscopy.
Applying techniques based on fluorescence spectroscopy, we performed a detailed thermodynamic and kinetic study to elucidate the Zn2+ mechanism to the C-terminal zinc finger of the nucleocapsid protein from HIV-1, NCp7 and of several of its mutants. We showed that Zn2+ binding and dissociation are multi-step processes, with the involvement of intermediates characterized by their protonation state, moreover the metal binding is connected to peptide folding. Fluorescence spectroscopy experiments in combination with NMR experiments and UV-Vis spectroscopy of the Co2+-substituted complex allowed to show that the highly folded and constrained structure of the C-terminal finger of NCp7 is related to the native Zn2+ tetrahedral coordination. The loss of the infectivity due the mutation of a single Zn2+ coordinating residue is not due to Zn2+ dissociation but to a perturbed conformation which prevents proper interaction with nucleic acids.
Fluorescence-based investigation of ligand binding to proteins.
Analyzing the interaction of NCp7 with various oligonucleotides with extensive fluorimetric titrations and fluorescence lifetime measurements, we showed a strong dependence of the binding parameters on the structure and the sequence of the oligonucleotides. To gain insight in the molecular mechanism of the interaction, it was greatly profitable to develop a method to simultaneously resolve the fluorescence decays of the two Trp residues of NCp7. This method can be also applied to other double-Trp containing proteins.
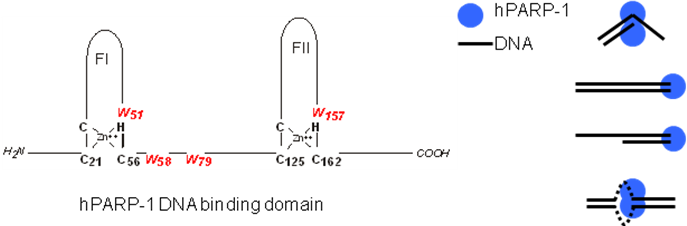
Furthermore, analyzing the DNA binding mechanism of poly(ADP-ribose)polymerase, hPARP-1, with fluorescence spectroscopy in combination with biochemical techniques, we revealed that the highly cooperative binding of two proteins is a prerequisite for activation and thus a rationale for molecular recognition. Binding of DNA by hPARP-1 is essentially independent of the DNA sequence; instead what is relevant for hPARP-1 functions is the recognition of overall features, like for example kinked structures.
Elisa Bombarda
